The ![]() Elliptec library is a lightweight Python toolkit for controlling Thorlabs Elliptec™ devices over serial links.
It wraps the sparse vendor examples into a clean API, handles multiple modules at once, and ships with a tiny CLI for quick bench tests. Neither the library, nor myself are in any way affiliated with Thorlabs.
Elliptec library is a lightweight Python toolkit for controlling Thorlabs Elliptec™ devices over serial links.
It wraps the sparse vendor examples into a clean API, handles multiple modules at once, and ships with a tiny CLI for quick bench tests. Neither the library, nor myself are in any way affiliated with Thorlabs.
I previously used it to automate optical experiments, and hoped to help others make it straightforward to integrate Elliptec stages into their own lab stack while prototyping new motion sequences. I no longer develop new features, and the code quality is science-grade at best, but once in a while I get an email or pull requests from someone who wants to escape the world of LabView.

Illustration of Thorlabs Elliptec modules.
In summer 2017 I took part in the 5th Summer School in Molecular Biophysics and Systems Biology 2017 and spent a month working on a project titled In-Silico Study of a Voltage-Sensitive Transmembrane Protein under Daniel Bonhenry, Ph.D. The project used the GROMACS molecular dynamics suite to model trans-membrane proteins sensitive to voltage differences above and below the membrane and studied the influence of various protein mutations on the behavior of the membrane.
Molecular dynamics simulations were carried out using GROMACS and the membranes were created using the Charmm-gui tool. The results were analyzed using VMD and partially automated by custom-writeen Tcl scripts. A more detailed analysis was performed using Python.

Computer simulation of a membrane.
During the summer of 2015 I returned to University of Puget Sound (link) in Tacoma, where I worked with Prof. Steven Neshyba (link) on further research of ice crystals.
Instead of viewing ice crystals in an SEM, I focused on computer reconstruction of the surface of ice crystals from already existing SEM images. Using four images captured by different detectors, we used the optimal estimation method to produce a 3D model of roughness occuring on the crystal facets.
In order to recunstruct the surface, we used CUDA, Python, MKL and the pyCUDA and scikit-cuda libraries. In further research, these 3D models can be used in scatter simulation to determine the impact of ice crystals in clouds on climate change.
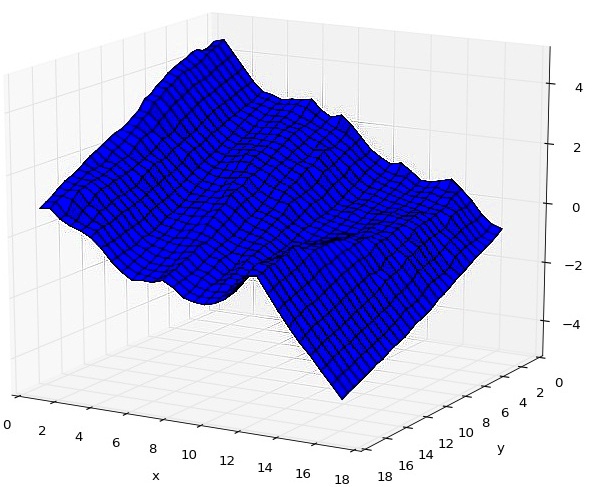
Reconstructed surface of an ice crystal.
Towards the end of sumer 2014, I was able to spend three weeks working at the Paul Scherrer Institute (link) in the northern parts of Switzerland. I focused on understanding the interactions of thin films of ice with na severu Švýcarska. Věnoval jsem se tam zkoumání interakce tenkých H2O2 in ari under the supervision of Dr. Thorsten Bartels-Rausch (link).
We studied the interaction by measuring the concentration of H2O2 in air before and after flowing through a thin tube covered with a thin film of ice on the inside. The amount of H2O2 binding to ice depends on the temperature of the air, the pressure in the tube, the method of growing the ice and on other parameters as well.
I analyzed the aquired data in MATLAB in
order to derive a precise amount of H2O2
adsorbed to the ice. A short summary of the project is available
![]() here.
here.

A part of our experimental setup.
During the summer of 2013 I was able to do research at the University of Puget Sound (link) in Tacoma, where I spent a month studying ice crystals using an SEM under supervision from Prof. Steven Neshyba (link).
Ice behaves differently depending on conditiond like pressure or temperature and creates various structures on a microscopic level. The main goal of this project was to grow ice crystals in the SEM chamber, focus on their facets and measure the roughness that forms on their surface.
Using this method, we studied how the statistical distribution of roughness depends on the conditions of
ice growth. This could lead to a 3D model of ice crystals at various conditions which could be used in
further research including scatter simulations. For more information, view the poster
presented on this topic
![]() here.
here.
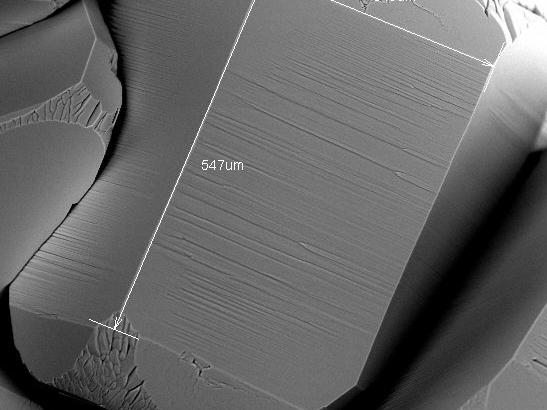
SEM image of an ice crystal.
During the summer of 2012, just before starting university, I participated in the summer school held in Nové Hrady by the Institute of Nanobiology and Structural Biology as a high-school student. In a four-week project, I modelled the interaction between Orosomucoid and several human hormones (warfarin, progesteron and propranolol) under supervision of Dr. David Řeha.
We used simulation package Glide from the Schrödinger Suite to model docking of hormones in the protein and looked for the best docking positions. We then used the Impact and Qsite programs together with Gaussian 03 to precisely calculate binding energies and re-prioritise the calculated positions. To double-check our results, we used molecular dynamics through the YASARA program.
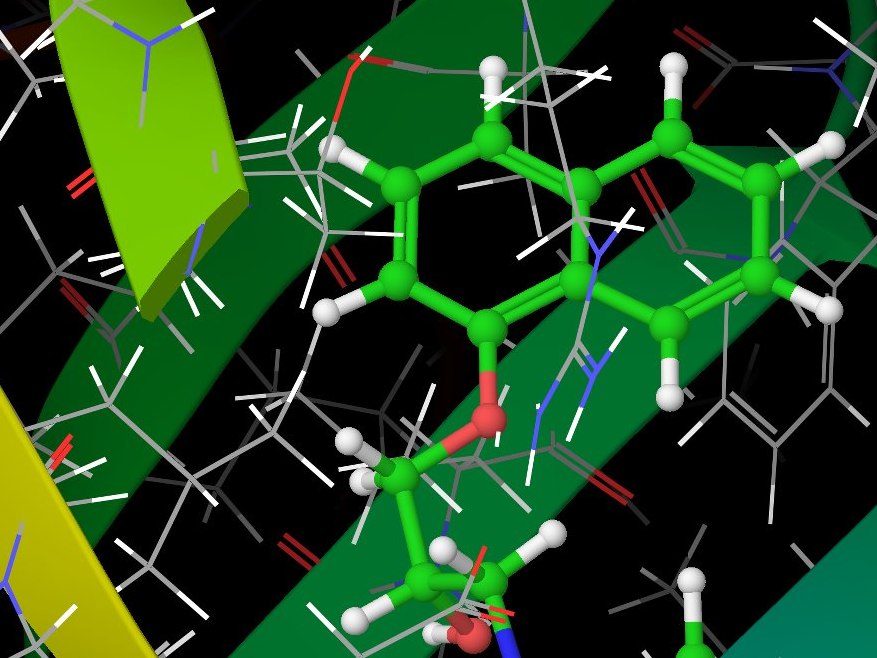
Computer simulation of Orosomucoid.